When creating a generator that parses an operational template definition (in OPT, webtemplate or other format) it would likely not be too hard to create both JSON schema and OpenAPI outputs in the process (they have a lot of similarities). Some tools like Altova Mapforce (and perhaps Mirth?) support Json schema and many code generators support openAPI.
A quick dirty hack POC version of schema+OpenAPI generator might be drummed up basing code on @bna’s web template → Asciidoc script that @ian.mcnicoll has continued on at https://github.com/freshehr/wt2doc (mentioned in post 20 above) and replace the DocBuilder.ts class etc. A better more properly model based converter may could likely be done fairly quickly by @borut.jures and his nice builder code or by something from @pieterbos & Co at Nedap or somebody else.
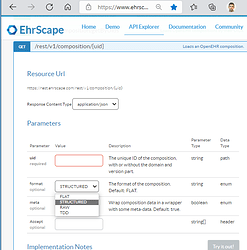
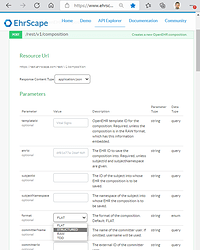
There could of course be many improved JSON structures to target, but to start with the simplified “structured” JSON that Better, EHRbase and others are using would be good to hav for integration tools that may want to loop over a structure. It’s called structSDT in Simplified Data Template (SDT) specification and COMPOSITION data instances can be both imported and exported in platforms like Ehrscape, see images below of the non-standardized way:
If using the standardised openEHR REST API I hope the same could be accomplished using the string application/openehr.wt.structured+json in the http-header Content-Type in the requests as described in the Alternative data formats section of the REST spec overview, when POSTing and GETting COMPOSITIONS. How many openEHR implementations support this already - ping @birger.haarbrandt @bna @pieterbos @borut.fabjan @stoffe @Seref and others?