At the SNOMED-meeting in Atlanta (I’m attending virtually ![]() ) we will hear about building FHIR questionnaires using the widget on the Nat’l Library of Medicine site. Nothing new, certainly, but as the CAP and ICCR cancer protocols are all now almost fully bound to their SNOMED-CT codes, it gives an exciting opportunity to see a functional algorithmic digital protocol with S-CT codes take shape.
) we will hear about building FHIR questionnaires using the widget on the Nat’l Library of Medicine site. Nothing new, certainly, but as the CAP and ICCR cancer protocols are all now almost fully bound to their SNOMED-CT codes, it gives an exciting opportunity to see a functional algorithmic digital protocol with S-CT codes take shape.
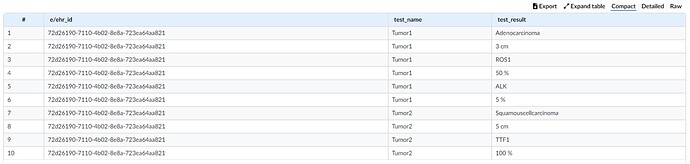
The widget is at https://lhcformbuilder.nlm.nih.gov/ . Click “start with existing form” and “import from local file”, which is the appended .json file for colorectal ca. Most of the work is by Alejandro Lopez Osornio and Hwang Ji Eun, a Korean postdoc working with Scott Campbell (University of Nebraska) at SNOMED International, I added some stuff, it’s dead easy. This is only a preliminary file shared a few weeks ago and work in progress, but since this came out I think Alejandro and Hwang have continued the build on colon and on other cancers, which they will present.
Colorectal-Cancer-STDU-v1.1.R4.json (42.2 KB)
I have as yet not found a way to extract the generated data into a report or summary, but from previous experience working with Sectra I know that that is not a difficult task - for the initiated! ![]() What appeals to me is the ease with which one can create a “goal” for a form in openEHR. I see it as a platform to easily add national or regional variables, grasp how it all functions and share a protocol with learned communities / pathologists at an early stage to get their input. I will pursue the matter and keep you posted.
What appeals to me is the ease with which one can create a “goal” for a form in openEHR. I see it as a platform to easily add national or regional variables, grasp how it all functions and share a protocol with learned communities / pathologists at an early stage to get their input. I will pursue the matter and keep you posted.
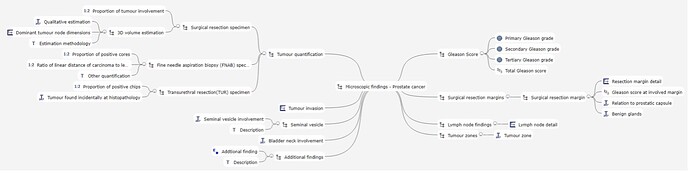
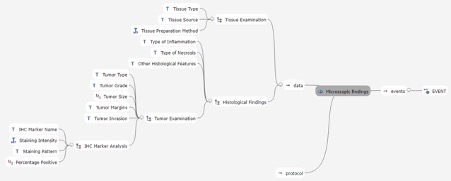
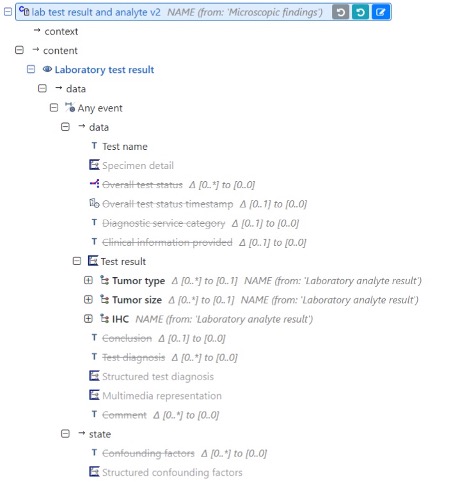
Meanwhile I’m working on the specifications files and MindMaps.
When have we planned our next meeting?