Hello everyone,
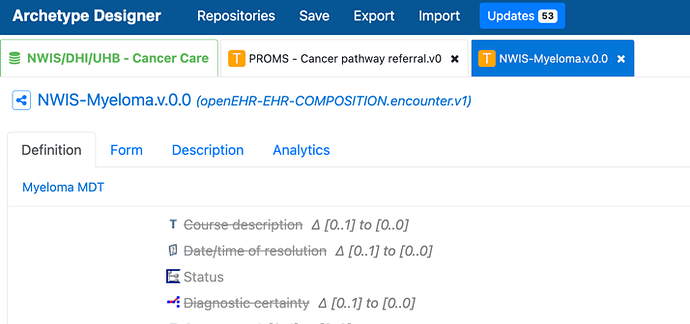
My name is Marlene, I’m fairly new to the community so I have tons of questions and no answers, but I would like to collaborate with your project @erik.sundvall. I am currently working in Wales where we are also trying to model our ‘Cancer Datasets’ for reporting and we have come across the same difficulty that you have.
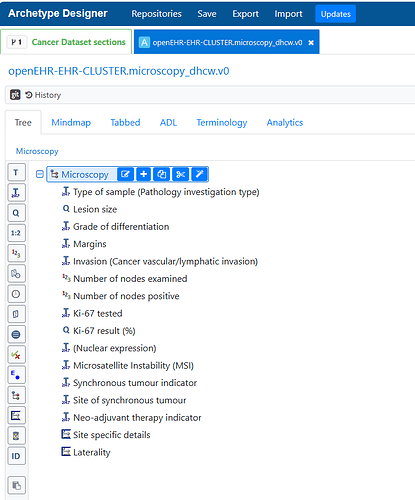
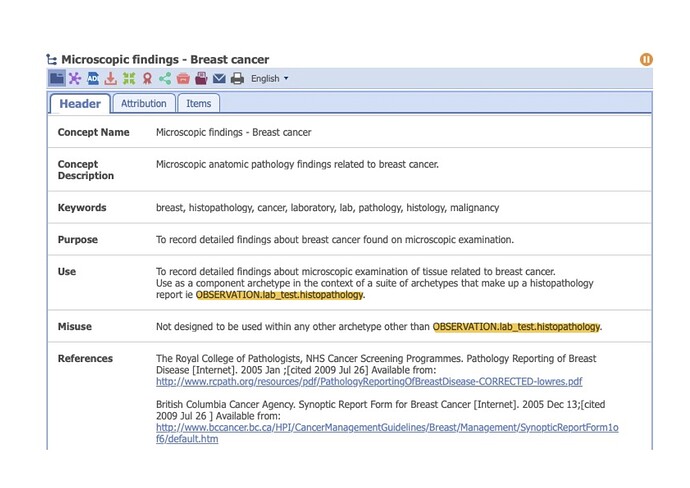
Currently, we have created a generic cluster archetype for microscopy details and we are trying to get it into the best shape possible but we know we re working based on a reduced number of cancers at the moment, and might need expansion. I don’t know how to share the archetype here to see if it could be a starting point.
I looked at the mind map, and I was wondering if you were planning on creating a specific cluster for each tumour site or build a generic and plug in smaller clusters with very specific details? As well, I see that you have included bilateral as a laterality, which isn’t in the openEHR laterality archetype. How are you handling that?
Out of what we have analysed, there are two points that we have identified as most challenging. First is the differentiation between solid tumours and haematological tumours as these have different pathology, location, staging, treatment. etc; the other are sarcomas, which can also be located anywhere and their classification varies. These also happen to be the most frequent tumours in childhood. Have you done any specific work on either of these?
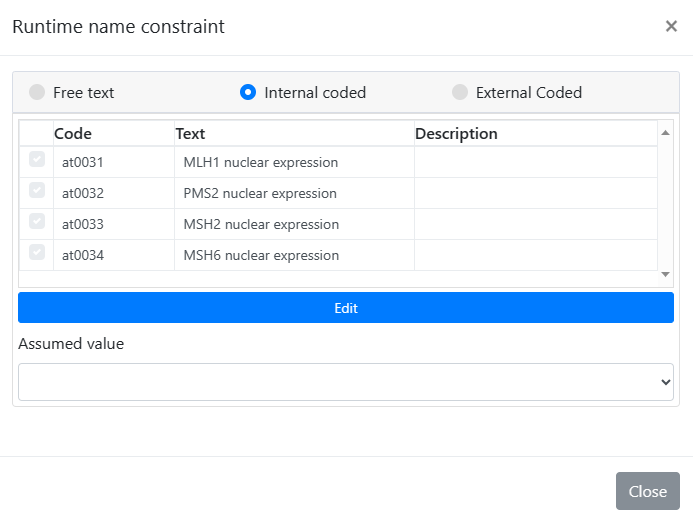
From a pathological point of view, would it make sense to have Elements for nucleic markers, cytoplasmatic markers, etc. to be able to reuse a generic cluster with whichever tumour? In our model we have used ‘runtime constrains’ for nuclear expression, but it might be ‘extendable’ for other uses.
As well, we are starting to do some work on tumour progression/metastasis (including lymph nodes)/transformation. Regarding this, it would be interesting to discuss if that ‘classification’ would need to be done at a higher-level archetype and then reuse the same microscopy clusters under that heading, or if it warrants a separate archetype to record how it was detected, when, etc.
Thanks for attaching: Clinical Knowledge Manager (openehr.org).
@ian.mcnicoll knows of our struggles  . It would be amazing to work on this together to align how we record this.
. It would be amazing to work on this together to align how we record this.
Thanks,
Marlene