Hi all,
I am a vascular surgeon interested in EHRs/EMRs. I am relatively new to OpenEHR. Is there a representation in the openEHR framework analogous to “OPCS procedure codes” or “HRG Codes” that can represent different surgical/radiological procedures?
Hi @sobath welcome!
I think the best entry point is the architecture overview document openEHR Architecture Overview
It touches all the basics.
Hi Sobath,
OPCS/ HRG are complimentary to what openEHR offers. Typically you would use a Procedure archetype, perhaps in an operative notes composition template, then the procedure name would be coded using an OPCS and/or HRG code.
The Procedure archetype is at Clinical Knowledge Manager
This is from the openOutcomes project
The template can be viewed at Archetype Designer
From a different use-case, her is how the data would look - where the procedure name is coded using SNOMED CT - it would be exactly the same for OPCS etc, and you can carry multiple codes if required.
"procedures_list/procedures/procedure:0/procedure_name|value": "Excision of palmar aponeurosis for Dupuytren's contracture of hand",
"procedures_list/procedures/procedure:0/procedure_name|terminology": "SNOMED-CT",
"procedures_list/procedures/procedure:0/procedure_name|code": "398211002",
"procedures_list/procedures/procedure:0/comment": "No intra-operative problems",
For imaging procedures we would normally record a result with
Dear @pablo,
Many thanks for your reply and the link. Please bear with me as I am trying to understand the basic concepts behind the openEHR framework.
Is there a need to try and represent every medical concept in archetypes? Since 50% of medical concerts become outdated in 5 years how do you keep up with the changes?
I believe you have to abstract the archetypes using templates and forms to manage the complexity for practical use at a busy clinic or hospital ward. But why do you need such complex modelling when clinicians will only use an abstract version of it?
With modern NLP-NER techniques do we actually need to represent all these concepts analogous to Entity–> relationships in an RDBMS?
What you are asking is why do we need information models?
The simple answer is that once you move out of a very specific, controlled context, it becomes very complex to understand the data that is collected, and you may be very surprised at the variability of data recording by clinicians.
Ultimately the data has to be coherent and queryable across many different care contexts, use-cases and organisations. To do that you need a coherent, ‘opinionated’ target for data entry - whether typed in or captured via NLP etc.
Just as a simple example - do we know if your OPCS-coded procedure code is in the context of a planned procedure, a performed procedure, or indeed not even in a patient context at all?
Dear @ian.mcnicoll,
Many thanks for the reply and the explanation with the links. I have been exploring surgical concepts using the clinical knowledge manager.
I feel that something as complex as an operation is represented in a very high level of abstraction compared to the concept of blood pressure.
If you try to map all surgical concepts to the detail of blood pressure, how do you keep up with the changes in each surgical speciality?
Typically we would plug in a procedure details cluster, specific to each speciality/ procedure.
This is from the openOutcomes Hip arthroplasty template
and is based on a form used for outcomes reporting.
If nothing else, this acts as a conversation space where we might try to rationalise the datapoints and related coded valuesets, captured very variably even within the orthopaedic knee surgery community.
Is this hard, yes, does it keep changing yes. but I don’t think NLP shortcuts the standardisation/rationalisation process, though it may be a great way to enter the data. You still need to get some degree of clinical consensus on what are the questions and the related acceptable answers and other data constraints
Dear Ian,
Many thanks for your reply.
I hope you don’t mind all the questions. I am trying to understand the basic concepts.
I admire the enormous work gone into the openEHR and the 30+ years of experience underpinning the framework.
My problem is operative procedures keep changing very rapidly over time. Who is going to capture all these changes? I have been working within my own speciality in vascular surgery to model the operative procedures and use a hybrid approach with NLP.
Even I am overwhelmed with the complexity and version control due to the rapid changes in my speciality. As an academic exercise modelling each concept is very scientific. But at the practical level in the implementation how do you keep up with the changes? eg. template editing → validation → changes to forms → end-user training etc.
Welcome to our world!! Even quite experienced clinical informaticians often fail to understand the overwhelming complexity, even within a single speciality.
We certainly do not have a perfect solution but I think the openEHR methodology (coupled with terminology use) gives us the best approach possible.
We do not have to structure everything, large chunks of what you have might reasonably stay as free text. Perhaps the NLP can be be applied downstream - that is fine for analytics where inaccuracy is tolerable but it is no good for operational use.
When things do need structured/codified, you need to be able to get real domain expertise (and that means debate and variance), and that’s where archetypes are really helpful in giving clinical domain experts a change to really pin down and agree what they can. And yes it it will change repeatedly, so this needs to be an ongoing conversation with versioning so that developers know what they are working with , as it changes.
The cool thing about CDR technology is that once any changes are made and agreed, they can be deployed without any further database analysis , entity modelling etc - essentially this is a no-code dataset deployment ecosystem.
And yes, that means changes to forms and training but you will have that regardless of the technology you use, and you are not compelled to change everything locally any time an international archetype is updated. It is up to your local implementation and clinical community to decide if/when to update.
Hi @sobath
It depends. In general an archetype represents information about a topic or concept. In some areas structured information is needed about the concept, so yes an archetype is needed. In other areas you just need to reference a concept, not to have structured data about it, in that context you just need terminology/vocabulary (a coding system like SNOMED CT).
When you need archetypes, there is an archetype lifecycle, since medicine and information requirements change in time, so do archetypes. They can be updated, versioned and even be obsolete and superseded by newer ones. The nice thing is: the information recorded complying with old archetypes is not lost, it just complies with a structure that previously (at the time that info was recorded) was totally valid. So you can still query and use that older info. In fact, one goal of openEHR is to have a lifelong clinical database, so your clinical information follows you through your life, and maybe after that (useful to your family).
Archetypes should try to be maximal data sets, but they can start small considering only current requirements, then be improved and completed later. Then with templates, built on top of a set of archetypes, you can manage the complexity, and make them as simple or complex as the context/requirements needs. From a template you can cherry-pick specific fields from archetypes and use only those, not the whole archetype structure. On the other hand, if the information you are trying to model and record is complex, the models will be as complex as that. Complexity of clinical information is another related topic. So sometimes you feel the openEHR models are complex, but is really because they model a complex reality (though to be correct those models are not modelling “reality” but the information that is generated by real life events or processes).
You will use NLP-NER techniques over free text, not over structured data. The focus of openEHR is structured data, though it can be used for narrative text records, coded text records and multimedia records. The goal of NLP-NER would be to get structured data from narrative text, then you can manage that structure with openEHR models. Mapping objects to a relational database (ORM) is a different thing, I don’t think it’s a good analogy here.
Dear @ian.mcnicoll and @pablo,
Many thanks for your clarifications and explanation.
As a use case; how does openEHR represent a bypass surgery eg. Common femoral to distal anterior tibial artery bypass using ipsilateral reversed great saphenous vein?
If I want to document that the pulse of the bypass graft behind the knee was palpable on my EHR what are the archetypes underpinning this documentation? Assuming that this data would be very important for reimbursement, quality assurance and research.
It depends if you want to record structured information about the procedure or if you want to identify the procedure. The first could be done by openEHR archetypes, the second is related to terminology not to structured information (see the comment about that in my previous message).
I want to do the former (structured information about the procedure ). So that we can perform queries to interrogate concepts related to operative procedures.
What archetypes do I need to use? I couldn’t find an archetype for bypass in the Knowledge Manager.
And how do you record the pulse of the bypass graft? The current pulse archetype is limited to the pulse of a native blood vessel.
I don’t know if there exist an archetype for that specific procedure. If not, it would be useful to document your requirements and ask clinical modelers, or propose an initial archetype yourself in the CKM.
Dear dr. Prem,
Welcome to the openEHR community, and thank you for your most interesting questions. I agree with Ian, openEHR is by far the best we have to get a chance of modelling the clinical world. And I agree with Pablo, openEHR archetypes are only ever as complex as the clinical concepts.
Regarding your statement 50% of clinical concepts are outdated in 5 years, I’m wondering what kind of concepts you are referring to? But even if you’re right, openEHR makes the technical side of modelling and implementation very easy, it’s just the understanding of the domains information aspects and requirements that will always be hard.
And the amount of work needed to spent on that can be nicely equivalent to the value of standardisation.
Given your example of palpable pulse behind the knee after bypass is quite interesting.
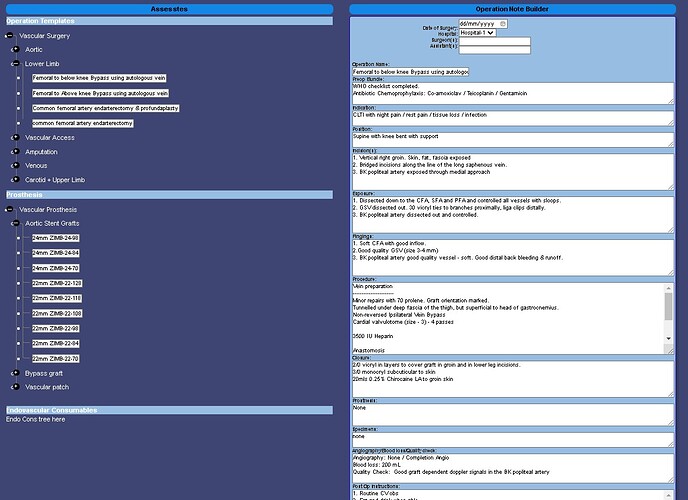
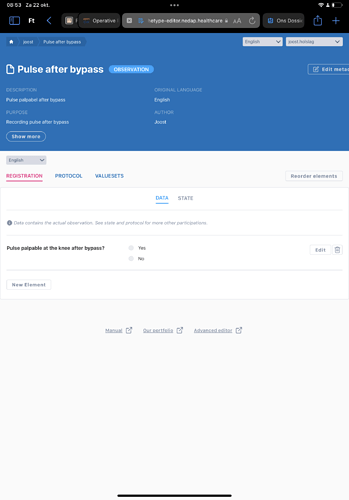
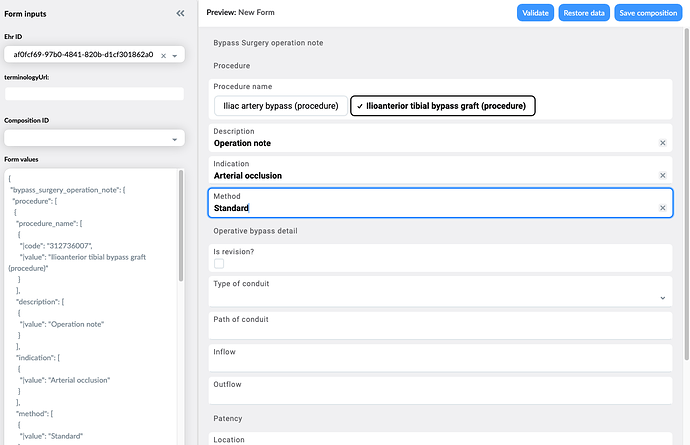
A quick and dirty model can be made into a form and deployed within 10 mins, see screenshot.
Now if you want to model this properly, there’s quite a challenge, and I don’t think the pattern has been figured out yet. Some thoughts:
The main concept may be the presence of a pulse. Which mostly fits with the pulse/heartbeat archetype. One could easily add a value to body site for “dorsal side of knee” and use snomed coding to standardise that value.
Now to model it’s a pulse measured after bypass graft, one could use the action.procedure with procedure name set and encoded using snomed again.and the pathway step set to completed.
Now if this measurement is done in context of a postoperative check one could use a template on composition.report-procedure.
Now there are multiple debatable points in this strategy. One way that is helpful to me to scope the concepts is to look at it from the querying perspective. If I query for “pulse/heartbeat” does it make sense to have this value returned. My best guess is “yes”, since a pulse in the graft quite certainly means there is a pulse and heartbeat. (Unless it’s still in a donor, or on a pump or so)
Dear Dr Joost,
Many thanks for your reply and the compliments. Regarding the first point;
Regarding your statement 50% of clinical concepts are outdated in 5 years, I’m wondering what kind of concepts you are referring to?
I am still unclear on the definition of a “concept” in the openEHR framework. But I can explain using an example.
The gold standard treatment for varicose veins ten years ago was vein-striping. Then came Sub-fascial endoscopic ligation, foam sclerotherapy, ultrasound-guided laser, ultrasound-guided radiofrequency ablation (current gold standard), ambulatory conservative hemodynamic treatment venous insufficiency (CHIVA), pharmaco-mechanical ablation, cyanoacrylate glue embolisation and Echotherapy over the last 10 years. This is just one surgical procedure. There is an exponential growth of techniques in modern surgical specialities as new techniques are adopted. How are the modellers going to keep up with all these changes?
For the second point, I feel that you will have to create a new concept (archetype?) for “bypass surgery”.
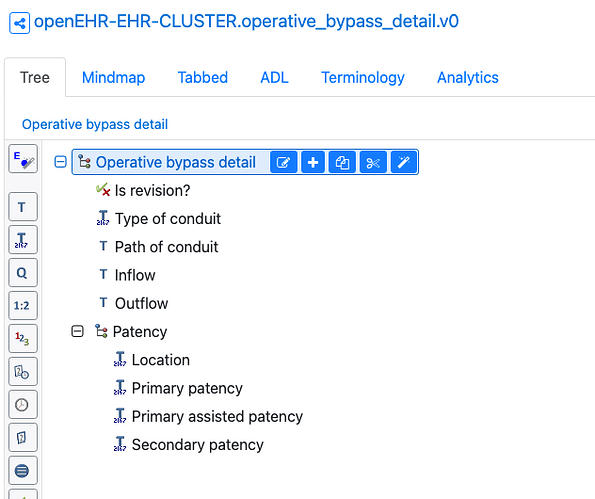
The pulse of the bypass conduit would be recorded at Bypass > Patency > Location of patency.
But bypass operations are performed by vascular, cardiothoracic, transplant, and bowel(upper GI, lower GI) surgeons hence more complexity will be added. I haven’t the knowledge or experience to properly model this concept but I am happy to help.
You are absolutely correct that this is a huge challenge, and of course, is equally true in almost every other specialty. However, it is not a challenge that every data/database/concept modeller faces, especially when within a speciality individual organisations, specialities and and even individuals may have somewhat different recording practice. And then each application developer has to acquire that knowledge from those clinical teams (again and again), and then turn that into computable artefacts. These are likely to be non-interoperable, so a different set of work then has to be undertaken to develop exchange models and related transform code to allow data exchange between the different siloed apps.
The openEHR premise is that we can save all of these application developers a lot of time and effort, if we try to agree the definitions from the outset, make that definition process and review very easy for clinical folks to engage with , and then have smart datastores that allow for immediate deployment of the models without further engineering.
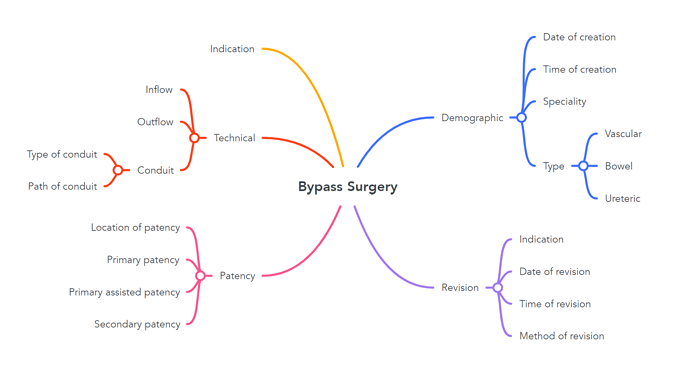
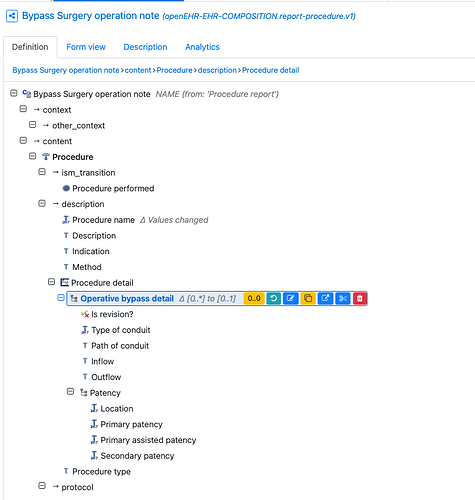
Nice start for a data model!!
Operative bypass detail archetype
And in context of a By pass surgery note 'composition and Action Procedure archetype.
The dates/times and context are handled by the Composition and Procedure.
And as @joostholslag pointed out in his version, these are immediately deployable to an openEHR CDR (datastore)
This is a crude form built from the template plus you can see the JSON data being built up at the left-hand side - this is what is sent to the CDR and is immediately stored and queryable.
Dear dr. Prem,
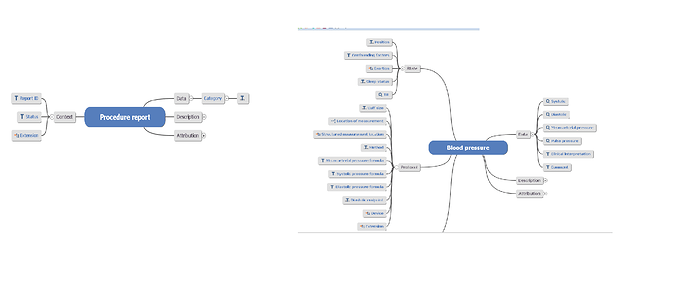
The key question in this interesting challenge of how to model “bypass surgery” is in defining “concepts”. The mindmap is a very helpful example of how one could define “bypass surgery”. It makes sense as a mental model for a surgeon to understand what a bypass surgery is.
Now in openEHR mapping out a concept is a useful and regular step when modelling a clinical concept like bypass surgery. But the perspective on defining a concept is oriented towards processing information regarding the concept not (necessarily) around understanding it.
This perspective change can be quite a challenge for us practically oriented doctors, but one I found very rewarding. So I hope I can interest you to follow one openEHR modellers perspective.
One starting question is what kind of data would you like to record around “bypass surgery”? This is nicely captured by the mind map you shared. Though other users probably want to record additional information regarding the bypass, for example suturing technique, intra-operative anti-coagulation used etc. etc.
A second key question is in what context (situation) this data will be recorded.
The openEHR view is that data entered into the system at one moment should be a complete “form”. This means, a report describing the procedure will have many of the elements described (date of creation, specialty, donor vessel etc.).
While others: e.g. “indication” may be recorded at a preceding event (clinic visit where the indication for surgery was determined).
And e.g. the patency and the improvement in (lower leg/foot) perfusion and reduction of symptoms will be determined after the procedure in a postoperative check.
So in openEHR this will probably be 3 separate forms. These “forms” are called templates.
Now there’s a question of what data elements have already been modelled. The CKM archetypes, especially the “published” ones are extremely high quality in my experience, so I would definitely recommend always using those. E.g. the action.procedure archetype already has an element “indication” that should be reused.
(Reuse of an element from an archetype is part of “templating”: where the template contains references to elements of archetypes and constrains them to be specific for the form. E.g. use the indication element, fix the procedure name to “fem-pop bypass” and don’t use the “urgency” element from action.procedure. and usually combining elements from many archetypes into a single template.)
Now even stronger reuse is recommend for data elements of the underlying openEHR reference model (RM). E.g. the “date and time of creation” and the “specialty” are elements already mostly defined in the RM.
So now the question is how to model the elements that are not yet part of an existing archetype. Here the key question is “what is the scope of an archetype”?.
One of my preferred ways to reason about this is to look at it from an information retrieval/querying perspective:
If I ask the database for information e.g. blood pressure it should give me all data regarding what is generally understood as blood pressure regardless of the form it was recorded in. (Systemic arterial Blood pressure is the same concept in a diabetes checkup form as in a “arterial insufficiency/claudicatio intermittens” exam, but a pressure in a toe in arterial disease is not).
So the quest is to find the underlying concepts of a bypass surgery that holds together along many different usecases and query’s. Getting this right is hard, getting it wrong is dangerous, once people/machines start to make conclusion from the data, e.g. using decision support algorithms.
This will require experience (good and bad). And is a process of continues learning and adaptation we go through as a community. And it’s one of the key reasons we hold public reviews of archetypes.
Luckily there are resources to help, e.g. described design patterns, asking experience modellers, [old review comments](h Clinical Knowledge Manager) in CKM, discourse discussions etc. etc.
The best modeller I know is @heather.leslie author of most of the CKM archetypes.
For modelling specific procedures, I’m not aware of any established detailed design patterns. The good thing is that once we have that it will be much easier to do additional procedures.
I agree with you and Ian that it’s a lot of work to model all procedures in the world.
However my hope is, a smallish group will be able to determine reusable design patterns using a basic library of core archetypes and reusing clinical terminology like snomed. Tools to do this modelling are also improving along the way.
And so I hope the work of modelling the bulk of the concepts (e.g, operatives procedures) can be done by many people with only some basic openEHR training. My utopia is that the original authors of a procedure, when publishing in a scientific journal also share an archetype modelling the procedure. Reusing this archetype in a live openEHR system then is a piece of cake, as I (partially) demonstrated in my example above.
So I do have good hopes that we have a system with openEHR that is up to the challenge you describe, of modelling all clinical concepts, even though they change rapidly.
Now if I made you eager to get started on modelling “bypass surgery”, I’m very curious for some details on your ‘project’: usecases, technical details (CDR), input forms, number of users/hospitals etc. Etc. ![]()
The key point here is that ‘someone’ has to do this and right now that is being done for every new procedure or method by thousands of different clinical application implementers right now without any coordination and where standardisation is an afterthought.
Part of the academic work to develop and document a new procedure or method should include a related data model from the outset. Clinical informatics input should be just as key an input to any paper as a statistician.