As I mentioned, this is in the spec: Base Types
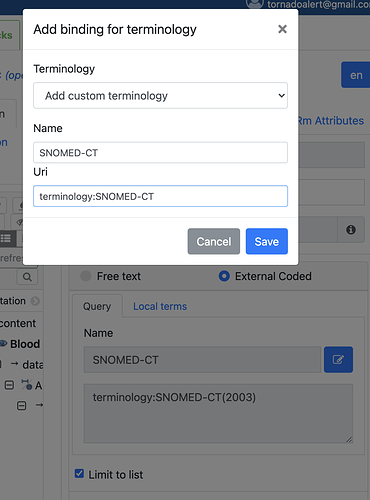
Check TERMINOLOGY_ID currently doesn’t accept “ICD10AM(1998)”
Not sure what you mean with the antlr grammar, we are referring to different things.
(* ------------------------- UID, OID, UUID -------------------------- *)
uid = iso_oid | uuid | internet_id ;
iso_oid = number, { '.', number } ;
uuid = hex-number, '-', hex-number, '-', hex-number, '-', hex-number, '-', hex-number ;
(* --------------------------- INTERNET_ID --------------------------- *)
(* According to IETF http://tools.ietf.org/html/rfc1034[RFC 1034] and *)
(* http://tools.ietf.org/html/rfc1035[RFC 1035], as clarified by *)
(* http://tools.ietf.org/html/rfc2181[RFC 2181] (section 11), *)
(* and relaxation of https://tools.ietf.org/html/rfc1123[RFC 1123] *)
(* The syntax of a domain name follows the grammar below. Slightly *)
(* reduced for the purpose here, plus allows underscores. *)
internet_id = subdomain ;
subdomain = label | subdomain, '.', label ;
label = alphanum | alphanum-ext-str, alphanum ;
(* -------------------- HIER_BASED_ID, UID_BASED_ID ------------------ *)
hier_object_id = uid_based_id ;
uid_based_id = root, [ '::', extension ] ;
root = uid ;
extension = ? any string ? ; (* any string *)
(* ------------------------- OBJECT_VERSION_ID ----------------------- *)
object_version_id = object_id, '::', creating_system_id, '::', version_tree_id ;
object_id = uid ;
creating_system_id = uid ;
(* ------------------------- VERSION_TREE_ID ------------------------- *)
version_tree_id = trunk_version, [ '.', branch_number, '.', branch_version ] ;
trunk_version = number ;
branch_number = number ;
branch_version = number ;
(* -------------------------- ARCHETYPE_ID --------------------------- *)
archetype_id = qualified_rm_entity, '.', domain_concept, '.', version_id ;
qualified-rm-entity = rm_originator, '-', rm_name, '-', rm_entity ;
rm-originator = alphanum-str ; (* id of org originating the RM on which this archetype is based *)
rm-name = alphanum-str ; (* id of the RM on which the archetype is based *)
rm-entity = alphanum-str ; (* ontological level in the RM *)
domain-concept = concept-name, { '-', specialisation } ;
concept-name = alphanum-str ;
specialisation = alphanum-str ;
version-id = 'v', ( '0' | non-zero-digit, [ number ] ) ; (* numeric version identifier *)
(* ------------------------- TERMINOLOGY_ID -------------------------- *)
terminology_id = name-str, [ '(', name-str, ')' ] ;
(* -------------------------- generic rules -------------------------- *)
alphanum = letter | digit ;
name-str = letter, { letter | digit | '_' | '-' | '/' | '+' } ;
alphanum-str = letter, { letter | digit | '_' } ;
alphanum-ext-str = letter, { letter | digit | '_' | '-' } ;
letter = 'A' | 'B' | 'C' | 'D' | 'E' | 'F' | 'G'
| 'H' | 'I' | 'J' | 'K' | 'L' | 'M' | 'N'
| 'O' | 'P' | 'Q' | 'R' | 'S' | 'T' | 'U'
| 'V' | 'W' | 'X' | 'Y' | 'Z' | 'a' | 'b'
| 'c' | 'd' | 'e' | 'f' | 'g' | 'h' | 'i'
| 'j' | 'k' | 'l' | 'm' | 'n' | 'o' | 'p'
| 'q' | 'r' | 's' | 't' | 'u' | 'v' | 'w'
| 'x' | 'y' | 'z' ;
number = digit, { digit } ;
hex-number = hex-digit, { hex-digit } ;
digit = '0' | non-zero-digit ;
non-zero-digit = '1' | '2' | '3' | '4' | '5' | '6' | '7'| '8' | '9' ;
hex-digit = digit | 'A' | 'B' | 'C' | 'D' | 'E' | 'F' | 'a' | 'b' | 'c' | 'd' | 'e' | 'f' ;
In RM 1.0.2 the syntax is a little different:
4.3.12.1 Identifier Syntax
The syntax of the value attribute is as follows:
-------- production rules --------
terminology_id: name [ ‘(’ version ‘)’ ]
name: V_NAME
version: V_VERSION
-------- lexical patterns --------
V_NAME: [a-zA-Z][a-zA-Z0-9_-/+]+
V_VERSION: [a-zA-Z0-9][a-zA-Z0-9_-/.]+
Note it allows XXX(123) and XXX(1.2.3), which the new syntax doesn’t allow.