Hi everyone. So this is my first post here, so please bear in mind that I may not know all the posts yet in this topic. I have a qustion regarding the polish HL7 CDA implementation of e.g. clinial document as OpenEHR file. The case is that, I am trying to remap Clinical document (like: Szablon 2.16.840.1.113883.3.4424.13.10.1.18 - plCdaDischargeSummary ) into template with proper archetypes. Could you give me any advice on that, I mean - how those values could look like in archetype designer? Thanks.
This is a template that mirrors the International Patient Summary not a CDR Discharge but should give a clue about what are some of the content detail archetypes you might want to use.
Remember too that the openEHR Composition class which is at the root of the template carries quite a few attributes that are targets for CDR attributes e.g CDA:Author → Composition/composer
Hi, in general CDA entries have one archetype each, and could be mapped to openEHR entries, structures or clusters, depending on the specific CDA concept.
Since CDA has 3 levels, depends on which level of CDA you have, how to better map it to an openEHR COMPOSITION (a ClinicalDocument in CDA is the equivalent of COMPOSITION in openEHR). Level 1 is CDA with non structured body, CDA level 2 is structured body with sections, and level 3 with entries and codes. In general a good mapping to openEHR is CDA level 3, which is the one that is more structured. Level 2 has sections with narrative text, and level 1 if for something like a PDF in the CDA.
Another thing you will find is the CDA header contains demographic data, which in an openEHR COMPOSITION you might not have, because the demographic data is linked to the EHR, and the EHR concept in CDA doesn’t exist. So in general what is on the CDA recordTarget is really an entry on an MPI and a reference from an openEHR EHR to that entry. That doesn’t mean you can’t put (technically) demographics in the COMPOSITION, is just that it’s not a good practice.
I’ve been working with both standards for a while and mapping is not so simple.
Hope that helps.
Thank you for that Ian and Pablo. In terms of the practical approach, could you give me advice on mapping constans? I mean…if there are slots in HL7 CDA PL like typeId (extension: POCD_HD000040 and root: 2.16.840.1.113883.1.3), then what kind of objects they are as archetypes in templates? Sorry for such basic questions, but I am very new in those EHR/HL7/FHIR standards.
Hi,
I’ll lash up a very simple partial composition template to give you an idea of the approach.
Ian
Thank you!
This is just a rough first cut based on the example data at Szablon 2.16.840.1.113883.3.4424.13.10.1.18 - plCdaDischargeSummary
As others have said, the patient details are out of scope and many of the Level 1 details are captured by openEHR Composition attributes that do not show in the Archetype Designer.
See Software Engineering Portal
e,g Composer is equivalent to CDA Author
Link to a Template Fileset that can be imported into Archetype Designer.
And finally Structured format Json example of an openEHR composition that matches the template.
{
"pl_-_cda_-_discharge_summary": {
"_uid": [
"37fd5e9a-ce9f-4998-9ba5-da61643f992e::example::1"
],
"context": [
{
"xds_metadata": [
{
"author_specialty": [
"Author specialty 64"
],
"class_code": [
"Class code 53"
],
"document_type": [
"Document type 51"
],
"confidentiality_code": [
"Confidentiality code 75"
],
"health_care_facility_type": [
"Health care facility type 59"
],
"practice_setting_code": [
"Practice setting code 40"
],
"event_code": [
"Event code 97"
],
}
],
"start_time": [
"2023-02-17T16:59:23.367Z"
],
"setting": [
{
"|code": "238",
"|value": "other care",
"|terminology": "openehr"
}
]
}
],
"encounters": [
{
"encounter": [
{
"encounter_date": [
"2023-02-17T16:59:23.367Z"
],
"category": [
"Category 71"
],
"event_type": [
"Event type 23"
],
"note": [
"Note 37"
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"encoding": [
{
"|code": "UTF-8",
"|terminology": "IANA_character-sets"
}
]
}
]
}
],
"diagnosis": [
{
"primary_diagnosis": [
{
"primary_diagnosis": [
"Primary diagnosis 17"
],
"details": [
{
"diagnostic_category": [
{
"|code": "at0064",
"|value": "Principal diagnosis",
"|terminology": "local"
}
]
}
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"encoding": [
{
"|code": "UTF-8",
"|terminology": "IANA_character-sets"
}
]
}
],
"secondary_diagnosis": [
"secondary_diagnosis": [
"Secondary diagnosis 61"
],
"details": [
{
"diagnostic_category": [
{
"|code": "at0066",
"|value": "Secondary diagnosis",
"|terminology": "local"
}
]
}
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"encoding": [
{
"|code": "UTF-8",
"|terminology": "IANA_character-sets"
}
]
}
]
}
],
"relevant_diagnostic_tests_laboratory_data": [
{
"research_results": [
{
"any_event": [
{
"test_name": [
"Reported results"
],
"media_file": [
{
"content": [
{
"": "http://med.tube.com/sample",
"|mediatype": "text/html",
}
],
"time": [
"2023-02-17T16:59:23.37Z"
]
}
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"encoding": [
{
"|code": "UTF-8",
"|terminology": "IANA_character-sets"
}
]
}
]
}
],
"pharmacotherapy_and_physical_rehabilitation": [
{
"epicrisis": [
{
"synopsis": [
"Synopsis 78"
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"encoding": [
{
"|code": "UTF-8",
"|terminology": "IANA_character-sets"
}
]
}
]
}
],
"prescriptions": [
{
"recommendation": [
{
"recommendation": [
"Recommendation 63"
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"encoding": [
{
"|code": "UTF-8",
"|terminology": "IANA_character-sets"
}
]
}
],
"medication_statement": [
{
"any_event": [
{
"manufactured_labelled_drug": [
"Manufactured labelled drug 69"
],
"time": [
"2023-02-17T17:04:26.799Z"
]
}
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"encoding": [
{
"|code": "UTF-8",
"|terminology": "IANA_character-sets"
}
]
}
]
}
],
"category": [
{
"|code": "433",
"|value": "event",
"|terminology": "openehr"
}
],
"language": [
{
"|code": "en",
"|terminology": "ISO_639-1"
}
],
"territory": [
{
"|code": "US",
"|terminology": "ISO_3166-1"
}
]
}
}
If I recall correctly this POCD_HD000040 is saying “this is a CDA in HL7 V3”.
If you need to map codes values, that would be the CD and other coded values in HL7 data types, that should be mapped to openEHR DV_CODED_TEXT.
But the important thing besides mapping data points is to map clinical concepts. For instance, when you read the implementation guide of the CDAs you need to map, check which clinical concepts are there and find the corresponding openEHR archetypes to map to.
Wow, thank you so much Ian, thank helped very much!
Pablo, my biggest concern was about coding constant values e.g. extension/root. Also, I need to dig more into Archetype Designer, as I had some trouble with generating the template from https://toolkit.cabolabs.com/ because there wasn’t any content in sample canonical json instance.
Remember that any TEXT or CODED_TEXT datatype can carry multiple code mappings, which includes putting mappings e.g to a CDA Section on the name/value attribute of an openEHR Section, as the name attribute (drawn from the LOCATABLE class is a DV_TEXT.
The only question is why and when you would want to do that. CDA is completely dependent on using external terminologies to label parts of the record. whereas openEHR uses archetype_node_id’s to do the same.
Practically this could be in STRUCTURED format
"diagnosis": [
{
" _name/value/_mapping" :[
{
"|match": "=",
"target|terminology": "LOINC",
"target|code": "29548-5",
}
],
}
],
"primary_diagnosis": [
"_name/value/_mapping" : [
{
"|match": "=",
"target|terminology": "SNOMED CT",
"target|code": "8319008",
}
],
{
I may not have those structures exactly correct but you’ll get the idea!
In canonical XML
<content xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:type="SECTION" archetype_node_id="openEHR-EHR-SECTION.adhoc.v1">
<name>
<value>Diagnosis</value>
<mapping>
<match>=</match>
<target>
<defining_code>
<terminology_id>
<value>LOINC</value>
</terminology_id>
<code_string>29548-5</code_string>
</defining_code>
<target/>
</mapping>
</name>
<archetype_details>
<archetype_id>
<value>openEHR-EHR-SECTION.adhoc.v1</value>
</archetype_id>
<rm_version>1.0.4</rm_version>
</archetype_details>
Thank you Ian for additional information. Now I have another question regarding submitting composition in flat format. As I have webtemplate, I can check the values to submit (please see on the attachment)
WebTemp.json (56.8 KB)
Also, the OPT file:
PL_CDA_DischargeSummary.opt (85.9 KB)
But regardless on value that I submit:
{
"ctx/language": "en",
"pl_cda_dischargesummary/composer|name": "en",
"ctx/territory": "GB",
"pl_cda_dischargesummary/context/cd_metadata/classcode|code": "DOCCLIN",
"pl_cda_dischargesummary/context/cd_metadata/classcode|value": "classCode"
}
my response is like:
"/context/other_context[at0001]/items[openEHR-EHR-CLUSTER.cd_metadata.v1]/items[at0030]: Attribute has 0 occurrences, but must be 1..1...
How could proper flat json submission look like for defined values like classcode?
Thank you
In some cases that is mapped to DV_CODED_TEXT, in other cases those could be UID_BASED_ID in openEHR. There is no magic there, you need to map code by code by it’s meaning.
The openEHR Toolkit doesn’t generate templates, you need to provide an operational template (OPT) and it will generate valid openEHR COMPOSITION instances based on the constraints of the OPT.
Can you publish the OPT you tested with? (I’m the creator of the openEHR Toolkit and might be able to help you there)
-
What CDR are you using and which API call?
-
Can you upload your example composition.
"pl_cda_dischargesummary/context/cd_metadata/classcode|code": "DOCCLIN",
"pl_cda_dischargesummary/context/cd_metadata/classcode|value": "classCode"
If you are sending CodedText you need to supply the teminologyID
"pl_cda_dischargesummary/context/cd_metadata/classcode|code": "DOCCLIN",
"pl_cda_dischargesummary/context/cd_metadata/classcode|value": "classCode"
"pl_cda_dischargesummary/context/cd_metadata/classcode|terminology": "LOINC"
Dear Pablo,
Here you have the OPT file:
PL_CDA_DischargeSummary (1).opt (142.2 KB)
Actually I came up with an idea how to do API request (sample below):
{
"ctx/language": "en",
"pl_cda_dischargesummary/composer|name": "en",
"ctx/territory": "GB",
"pl_cda_dischargesummary/context/cd_metadata/classcode": "DOCCLIN",
"pl_cda_dischargesummary/context/cd_metadata/id/root": "EVN",
"pl_cda_dischargesummary/context/cd_metadata/id/extension": "DOCCLIN",
"pl_cda_dischargesummary/context/cd_metadata/title": "DOCCLIN",
"pl_cda_dischargesummary/context/cd_metadata/effectivetime": "2020-06-08T17:04:30.621Z",
"pl_cda_dischargesummary/context/cd_metadata/confidentiality_code/code": "A",
"pl_cda_dischargesummary/context/cd_metadata/confidentiality_code/codesystem": "2.16.840.1.113883.5.25",
"pl_cda_dischargesummary/context/cd_metadata/languagecode": "test",
"pl_cda_dischargesummary/context/cd_metadata/setid/root": "test",
"pl_cda_dischargesummary/context/cd_metadata/setid/extension": "test",
"pl_cda_dischargesummary/context/cd_metadata/versionnumber": "test"
}
For now I have no other issues regarding making the requests, only to provide detailed template. On that topic, I’ve encountered a problem - for the internal/external coded text - is there possibility to provide csv list of objects? I have a few hundreds values for race: https://www.cez.gov.pl/HL7POL-1.3.1.2/plcda-1.3.1.2/plcda-html-1.3.1.2/plcda-html-1.3.1.2/voc-2.16.840.1.113883.1.11.14914-2014-03-26T000000.html and filling the table by hand could take pretty long…
Is there maybe a terminology service available with a FHIR API? Depending on your openEHR server, there is direct support for external value sets (EHRbase and Better Platform support this for example). You can find some examples for the correct binding within templates like here:
The CKM resolves the reference against a central FHIR-Terminology Server instance.
Edit: the documentation for EHRbase may give you some better idea how to use it (8. Terminology Validation — EHRbase documentation)
Thank you Birger for that. I have found the option for changing the terminology server and there is most of that what I need.
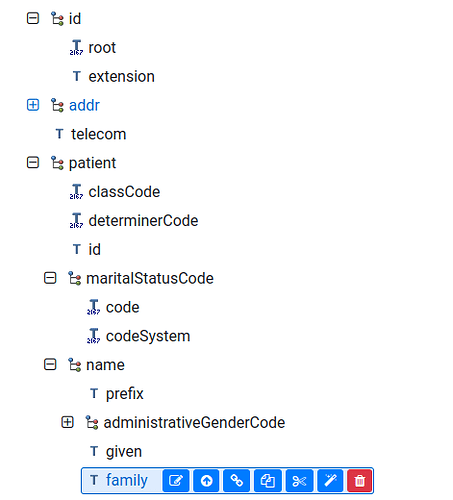
About the uploading composition, I have some issue. On the picture below there is structure of my template (from archetype):
But when I am trying to upload the comp. there is an error for name object. Part of my flat json is:
"pl_cda_dischargesummary/plcdabaserecordtarget/dane_pacjenta/recordtarget/patient/name/given:0": "PSN"
And this is the error that I have:
Could not consume Parts [pl_cda_dischargesummary/plcdabaserecordtarget/dane_pacjenta/recordtarget/patient/name/given:0]
I know that for objects with cardinality
0..*/1...*
(as id and name)
I must give an index in there, but it is not working anyway. As an attachment I send the OPT file.
PL_CDA_DischargeSummary.opt (232.7 KB)
However, for id it is working (as below):
"pl_cda_dischargesummary/plcdabaserecordtarget/dane_pacjenta/recordtarget/id:0/root": "2.16.840.1.113883.3.4424.1.1.616",
"pl_cda_dischargesummary/plcdabaserecordtarget/dane_pacjenta/recordtarget/id:0/extension": "fsdf",
Is there anything that I am missing something there?
Thank you