Pardon my boldness, but I tend to agree with @thomas.beale’s point, even as a clinician.
It’s also obvious, from this discussion, that clinical modellers aren’t happy with the way specialisations work, at least in ADL 1.4.
Ontologies confer meaning, and are one of openEHR greatest strenghts (e.g. archetype classes, naming, paths, etc). Although I understand the fear of ‘over-ontologizing’, we all feel the pain of ‘under-doing-it’.
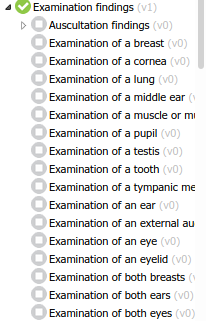
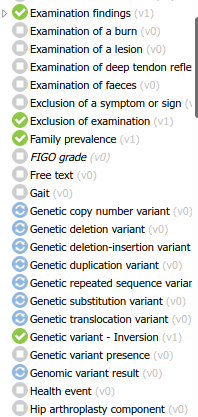
After opening and taking screenshots of all 547 openEHR artifact mindmaps in International Community’s and Apperta’s CKMs (for the sake of quick browsing), I must say the repositories feel a bit ‘flat’ inside each class.
Besides nesting (almost) all exam findings as such, the archetype list is a bit hard to navigate. For the least acquainted, it gets quite hard to find what you are looking for. It’s even harder to be sure there is not a structure defining what you need - which is a common use case for us, and probably for all modellers.
It’s curious that our team discussed this topic exactly after noticing the different pattern of archetype governance in those two (somehow) similar modelling subjects: physical exam and genetics. In both scenarios, the main goal is to describe a reality, pointing out specially what doesn’t conform to the ‘normal’ pattern. We settled in the opinion that, in the second case, the goal was just to identify the anomaly, not to describe the full genome (or even a part of it).
Besides allowing for intelligible repositories, this seems like a good policy ‘tech wise’, for the sake of querying data or retrieval of task planning elements in each instance of a work plan.
So, wrapping up, it feels like ‘is_a’ relationships should be part of the metadata of archetypes - and the ‘semantic slot’ could be very helpful in some use cases. In what measure that depends on specialising archetypes? I really have no idea.
PS: if anyone is interested in the images I mentioned, I’ll gladly share it with you