System: EHRbase
Version: v0.32.0
I’ve come across two problems that probably are closely related. I was doing some tests and I created a single archetype containing the full structure I needed. That is, a COMPOSITION, which includes an OBSERVATION, which includes several ELEMENT.
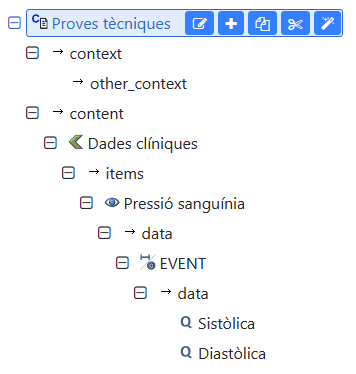
I know that’s not the common good modeling practice, but it is completely legal.
With that archetype I created the template, and then some instances using the Cabolabs openEHR toolkit.
To my surprise, when I tried to load those instances in EHRbase, I got this error (at0004 is the OBSERVATION):
{
"error": "Unprocessable Entity",
"message": "/content[at0002, 2]/items[at0004, 1]: Invariant Is_archetypeRoot failed on type OBSERVATION"
}
My deduction is that since the archetype_node_id in the OBSERVATION instance has an atNNNN code and not an archetypeID, it is thus rejected. I can understand that this kind of instances could impact in the indexing and querying processes, but nothing in the specifications says that it is an incorrect instance.
This deduction was supported later, when I was trying to do an AQL that returns all ELEMENTs in all instances.
SELECT ele FROM EHR e CONTAINS COMPOSITION c CONTAINS OBSERVATION o CONTAINS ELEMENT ele
First I got no results. Then I made another test creating an ELEMENT archetype, inserted it into the OBSERVATION, created instances, and only those identified elements were successfully returned with the previous query. That means that an archetypeID is needed to index and query data, although ELEMENTS without it are accepted by the server without complaints. I see there a lack of coherence:
- I cannot store OBSERVATION instances (I guess that it will happen with any kind of ENTRY) if they are not defined as an independent archetype.
- I cannot query ELEMENT nodes unless they are defined in an independent archetype, although they can be stored being “anonymous” (as it is always the case).
I insist, I completely understand that there could be efficiency reasons for these limitations, but they are affecting valid use cases of the specifications.
Any thoughts about this?