Hi all, late to the DICOM imaging party but letting you all know that I’ve been working on updating the imaging archetype family over a period of time.
The project is here: Imaging examinations project
I’ve just uploaded the latest version of the OBSERVATION.imaging_exam_result. In fact, you will note that it is still on a branch (so you can have a sneak preview while it waits for a NO translation to be added. We intend to send it out for review very soon. The design may appear a little different to the discussion in this thread and I’d like to explain why and then encourage everyone to join the review by adopting the archetype.
The focus of the Imaging exam result archetype is to record the findings and interpretation as part of a published result report. The current design is a (hopefully relevant and meaningful) mashup of:
- Clinical reporting standards/recommendations for imaging reports
- FHIR Diagnostic Report and Imaging Study resources
- DICOM attributes relevant to a report
- Alignment with the OBS for lab test result where relevant.
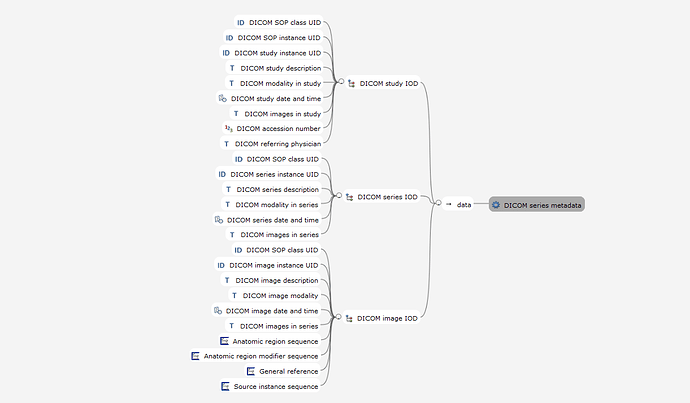
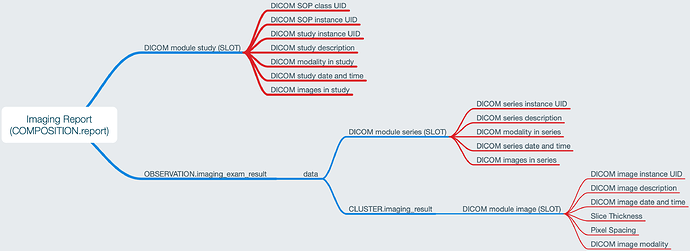
So I’ve designed the OBSERVATION to be for a single Study, potentially including multiple series which is modelled by a CLUSTER.imaging_series that can be used within SLOTS for the study report and to represent other series that may be used for Comparison purposes.
I had previously modelled a CLUSTER that carried both DICOM study and DICOM series level attributes but it ended up duplicating many data fields that were required in the result for including non-DICOM reporting. So we needed a different approach - today I’ve done a major redesign to include the Study level attributes within the OBSERVATION and the Series level attributes are kept in the optional CLUSTER.imaging_series which can be nested within the OBS at ‘Series details’ SLOT and ‘Comparison series details’ SLOT as required. You can see the changes if you view previous revisions in the same CKM branch.
In my investigation I’ve not come up with evidence that we need to document down to instance level for reporting purposes, but happy to be corrected. I very deliberately want to avoid replicate an RIS level of DICOM detail within an imaging report, only what is required for our purposes.
This is the art of archetyping in action, determining how far we need to incorporate DICOM and FHIR attributes but keep the archetype focused on the reporting purpose. That doesn’t mean that they are not required in other contexts and so we need to understand the other opportunities before we go much further.
You will also note a growing family of specialised CLUSTER archetypes that record the specifics of imaging exams of a growing number of body regions and associated metrics such as fetal biometry. It is anticipated that these will grow in number and level of detail over time. This imaging exam detail pattern is closely aligned with the way we are modelling physical examination.
@christian.haux et al - I’m curious to get your feedback and how these archetypes meet or miss your modelling requirements. I referenced the specialisations you pointed us to further up in the thread, and included most of them in my newest iteration but didn’t have a use case for a number of your attributes at present. So if you have use cases for the others that we need to add, please let me know
Regards
Heather