Hi all,
I’ve obtained a text from a data analyst, claiming the task of extracting data from openEHR format to serve in a data warehouse is extremely resource demanding and challenging. I’ll post it below, with some alterations not to disclose its author, nor the hospital or vendor in question.
If some of you out there with experience in analysis on openEHR data can provide examples of how to ease the burden or other comments to the text, I’ll be most grateful. In nice language, please ![]()
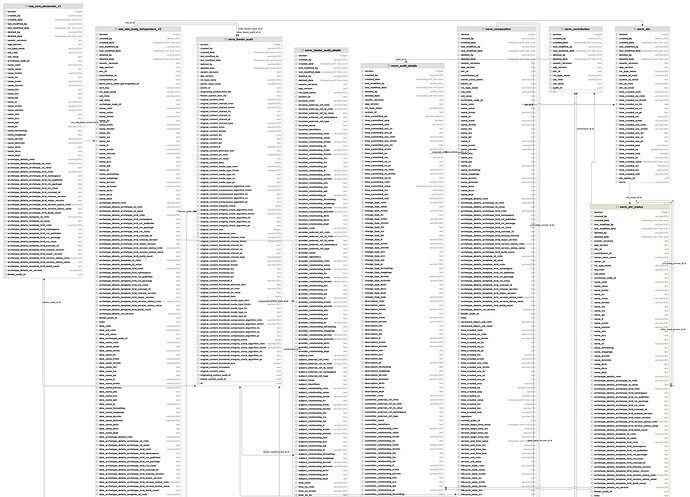
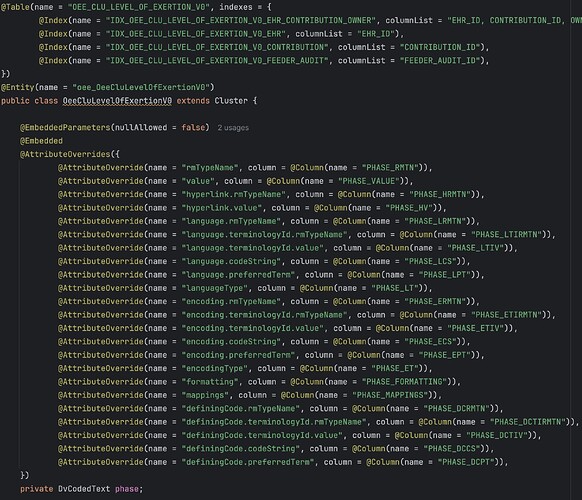
The introduction of a structured EHR has the potential to address several challenges arising from the lack of structured data in primary EHR sources at (a named hospital). However, the current archetype-based construction of (a named EHR product) (openEHR) presents multiple challenges, including the inability to use this data directly for analysis. The implementation of openEHR in the system makes it impossible to query the data for analytic purposes. The current solution relies on consultancy services and software from (a named vendor) to query and replicate the data so that it is suitable for analysis purposes ensuring acceptable response times even for high-volume querying. This results in a sole dependency on consultancy services from the vendor to access the structured data entered by the hospitals employees, and necessitates a robust plan for maintaining structured views when there are changes to templates/forms or archetypes
…
The extraction (of data from openEHR format to normalized relational database structures) is expected to be significant and demanding, requiring sufficient allocation of resources, both technical resources and expertise in openEHR/archetypes.
Comments, anyone? Pinging @birger.haarbrandt @ian.mcnicoll @yampeku