Hi David,
It’s difficult to provide a definitive response to such a complex modelling question via this forum. You’re working in a space that, while we’ve tried to anticipate it in the design of existing archetype patterns, remains relatively unexplored in practice. I don’t yet have direct modelling experience with this level of anatomical and physiological detail, and I can’t know what I don’t yet know.
To provide a thoughtful and appropriate response really requires a deep understanding of the context and specific requirements. Ideally, that would be through a face-to-face workshop over the course of a day, where domain experts are directly interacting with modellers. Translating language adds another layer of complexity that makes it even harder to give immediate or accurate feedback.
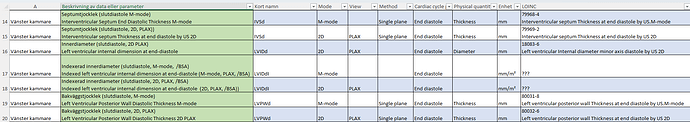
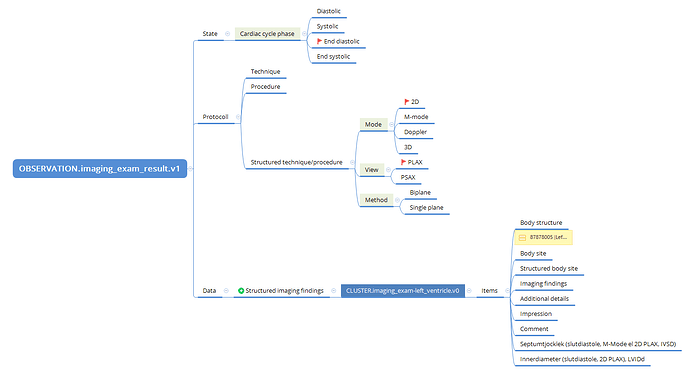
From first principles, I can say that we don’t typically create specialisations for left and right sides of a structure separately. So, if we need an archetype for imaging examination of a ventricle, I’d expect only one archetype, with the laterality (left or right) represented using the ‘Body structure’ element. This is consistent with how we approach other paired anatomical regions.
To do complex modelling justice, even with a solid understanding of the clinical background, it is common to enter a significant phase of exploration. There is usually a period of trial, error and iteration before the optimal approach becomes clear. In Australia, I often describe this as “going down a deep rabbit hole” and hoping to eventually find my way out. I’m not sure if that analogy translates well, but it feels apt. This is a normal part of my process, even with extensive experience. Each new clinical use case presents its own conundrums and we rely on existing patterns to support solving the next one. But your situation has little existing for us to leverage at this point in time.
At this stage, I do think it would be valuable to have guidance from senior modellers or CKAs to ensure your work aligns with CKM modelling patterns and has the potential to contribute meaningfully to the shared archetype library. Unfortunately, we don’t yet have the resources in place to fully support that kind of mentoring or review. I wish it were different.
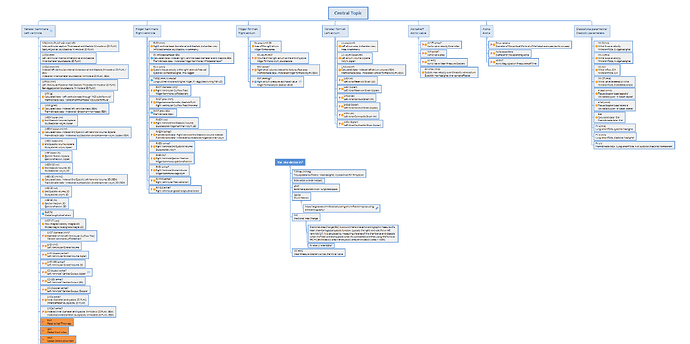
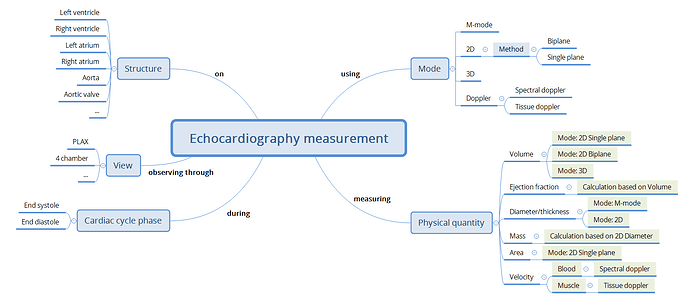
As a first step, perhaps consider collaborating around and sharing a mind map or structured outline that captures what data you would expect to record for each specialisation or structure. This might help clarify why a separate archetype may be justified, or whether your needs can be addressed through templating or compositional approaches. The complexity of phases of the heart is another layer on top of the basic identification of CLUSTERs required to correctly represent the structures that need to be imaged.
I hope this helps you make some progress. The more collaborative your approach, the more likely it is that the modelling requirements will become clearer. From there, we can hopefully work out a way to support translation into the correct scope for each of the archetypes that follow the established patterns.
This is important work, and thank you for your efforts.
Kind regards,
Heather