Hello, I’m looking to create a small Clinical Data Repository (CDR) that can handle template storage, composition storage, composition validation, and execute AQL queries based on these compositions. Up to this point, I’ve successfully implemented all aspects except the AQL functionality. I’ve been utilizing the EHRbase Maven SDK (org.ehrbase) throughout this process. Could you assist me in understanding how to transform an AQL query into an SQL select statement? Are there any examples available for this? I’ve reviewed the EHRbase codebase, but it appears they haven’t leveraged the SDK for this and instead have implemented a more intricate solution. Thank you for your help!
AFAIK what you ask for can’t be done. For generating any kind of SQL you need to know your relational schema. EHRBase has it’s own schema and is not 100% relational, it uses document type from Postgres mixed with relational. Then, if you have a different schema, the SQL+no SQL generated by EHRBase won’t work on your schema, and won’t work if you use something different than Postgres as DBMS. So you’ll need to create your own AQL processor.
best,
Pablo
Hi Pablo, thank you very much for getting back to me. I’m aware that you’re the primary developer and maintainer of the EHRServer. I’ve also taken the time to go through the source code of your project. From what I understand, it seems AQL hasn’t been implemented in your project yet. Could you please shed some light on the data query mechanism that EHRServer employs?
Hi Ali, yes I’m, though I also worked in EHRBase when they were migrating the code from EtherCis GitHub - ethercis/ethercis: START HERE: Documentation, internals and installation material, support info etc. Please post ISSUES here as well, thanks!
The challenges of AQL are many. One is that it requires some non-relational features since it’s basically a hierarchical query language, and most databases (relational or document) rely on their own query language to query over hierarchical data, so the final queries are (in general) not portable between different DBMS. Like EHRBase uses JSON structures internally, or some other implementations use a mix of relational and Lucene indexed data.
As mentioned before, those queries also depend on the schema, so if your schema on Postgres is different than your schema on MySQL or SQLServer, then queries are not portable between implementations.
Another challenge of AQL is that the spec doesn’t specify the whole language, it focuses on the syntax, but not really on what happens under it. I think the specs kind of relies on the reader to know the semantics of the keywords from SQL. So implementations could differ and you might get different data from different vendors, even if the same data was loaded and the same AQL query was executed. IMHO AQL is underspecified and we should work more on the querying model than on the syntax, so different implementations behave in the same way.
On a side note: as part of the SEC I worked on the AQL spec for about a year while working with EHRBase and that work was applied for many major improvements to the spec, though I think it’s not ideal yet.
My conclusion is that you can’t just take an AQL implementation and make it work over any database, though that would be great. Imagine having just one reference implementation for each major language, devs can just pick one and integrate it in their systems. That would enable innovation at many levels. Sadly we are far from that.
There could be some light on this by using recursive queries that was added to SQL and implemented by some well known relational DBs, that is Common Table Expressions. Though I don’t have time to start researching that area anytime soon. I believe with that we could improve query portability and the AQL spec in general, by providing the mapping to SQL in the spec itself.
For some of those reasons I decided to go in a different direction with querying on the EHRServer and Atomik implementations. I have designed a query model (it’s a model not a syntax) called Simple Archetype Query Model that translates into plain SQL so it’s portable between MySQL, Postgres, SQLServer, Oracle, etc. and it’s not a general query language, it constraints what and how you search, but it’s pretty flexible for doing most common things. We have a query builder embedded in EHRServer and Atomik, which allows you to create the queries visually and test them live. It also supports mixing SNOMED CT expressions in the queries for semantic querying.
There are some demos here https://www.youtube.com/watch?v=zSftiFBjboE&list=PL-4c1WHznyulrf8wPhaOq7T0E2QWQMCHH
That has been the elephant in the room for many years ![]()
What is being stated here?
If it is that different CDRs/AQL implementations should return the same results (for the same data), I assumed this has been true. I base this mostly on the openEHR messaging and on the incidental exceptions where ehrbase and better platform return different results. I know conformance testing needs to be much more elaborate to better guarantee this.
This being true (‘enough’) is crucial to openEHR, for clinical safety, application portability and other promise of the benefits of openEHR.
But if it’s about translating AQL to SQL so that different CDRs that do all use SQL can reuse code, that’s definitely nice to have, but not essential to the openEHR benefits.
It’s long/complex to explain, you’ll need to study the whole AQL and we’ll need to do a workshop or two to go through all the issues in the spec. Though I did study the specs and tried to improve what I could while working for HiGHmed, but don’t trust me, you can check the spec yourself.
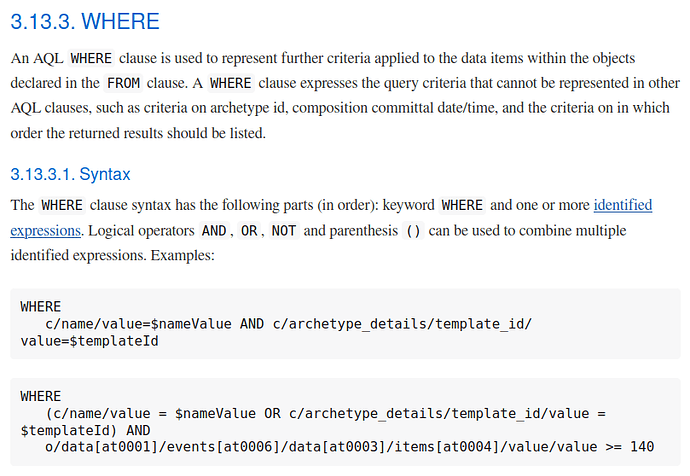
One small example, consider the WHERE clause definition: Archetype Query Language (AQL)
Then think about how that should be evaluated and executed and how that affects the results of a query, and all the issues that could happen from that evaluation/execution. You will find many things are not defined by the spec, for instance what happens when you have o/some/path > 123 and o/some/path points to a string, we don’t have a strongly typed syntax right now. The spec is focused on the syntax, not on the model and functions under it, and there is a good reason for that: when that is specified, it needs a reference schema, and since openEHR persistence is open for implementation, each vendor can define it’s own schema, so each vendor can implement AQL over different schemas in different was, then we have divergent results in some cases.
On the other hand, consider the WHERE definition in SQL:
Do you see the difference? SQL has a more mathematical/scientific approach for the definition. It also mentions how it applies to results (rows) in relation to the FROM clause, and gives some rules that defines how it should be evaluated/executed. All that is defined over a given model/schema that defines tables, columns, rows, etc. which we don’t have in openEHR.
Another point: AQLto SQL is not always the transformation you want. In fact a CDR could be implemented on a JSON or XML database, or on a graph database, so AQL will need to be translated to the speicific query language used by the database, like XQuery in XML dbs. For good or bad AQL resembles SQL which for newcomers might be confusing since most think that SHOULD be translated to SQL.
Certainly is not about that, it’s about all CDRs interpreting an AQL in the same way and execute exactly the same query (semantically) in the database brand they use, even if it’s no-SQL. That is why (for me) the model behind the query and the execution steps are more important than the syntax of the query. If two CDRs use the same query model and follow the same query execution rules, they can use any syntax they want to represent their queries as strings.
With that being said, I don’t think it’s practically possible to change the current spec in that direction, because it’s really difficult to define the DB schema, which is needed to specify the query model and execution details. Though I believe it’s possible to define some abstract/virtual model to have a framework that allow us to define those aspects in the spec, and that allows each vendor to map to their own technology stack. I also think this is a green area for innovation, since we could have different query languages for specific domains like data analysis, reporting, CDS, etc). For instance our query model is focused more on those areas than on being a general purpose query model (AQL is a general purpose query SYNTAX).
Hope that helps for the discussion.
Quick comment from the sideline (would deserve a more thorough reply but this is for later): I think we are not too far off between implementations as SQL semantics are implicitly expected. Of course there is lots of room for improvement, but we have done quite some testing between Better and EHRbase in HiGHmed and found differences not to be more severe as for example between different SQL dialects like Postgres, SQL Server and Oracle.
So, surely a bit more work to do on specs and conformance but reality is not as bad as it could be.
Thank you all for your insights. specially @pablo for his detailed explanation.
let me refine my question. Is it feasible for us to create a small application capable of storing and querying(with AQL) compositions? From your Pablo’s feedback, it seems the answer is no(or at least not straightforward). It appears the query processing logic isn’t available as a separate library, and the current AQL query processor logic of Ehrbase is complex and, to be honest, somewhat unstable.
Additionally, it relies heavily on PostgreSQL-specific functionalities. This issue extends beyond just dialogue; Ehrbase uses many stored procedures, complicating any migration process. I think Ehrbase is overly extensive for our needs, and its internal complexities hinder integration with other systems.
@pablo I have’t watch the video yet but does your query model supports aggregations and joining.
Some fair points, but be aware that the old AQL engine of EHRbase is about to be replaced with a new one which addresses your concern regarding stability. This being said (and while using way less database-specific functions in the new version), we are quite happy with the Postgres API and never had any requests to support other databases like Oracle or SQL Server. So there is no good case for us to make to make it agnostic of Postgres, so we prefer to rather embrace the ecosystem with lots of Postgres derivates likes YugabyteDB.
I would still be very interested to learn about the challenges for the migration process based on the db model. Given your project scope seems small to mid-size, I would expect that you can just use the openEHR APIs and you won’t have to deal too much with any database internals.
Hi Birger, thanks your your input.
I’m don’t have hard data on the size of the gap between implementations. Cross-verification is useful, but you still don’t know what is happening under the hood and which mix of clauses could lead to different results. The problem is not the implementations, which are fine pieces of engineering created by very smart people to try to do what the spec says, but the problem is at the spec level. With the ideal spec, you wouldn’t need to do cross-checking between implementations, just conformance checking of each implementation against the specs.
For anyone coming to openEHR, we should be clear about those issues and challenges. AQL as a spec has its value, that is undeniable, but also has its shortcomings. Though we could argue that is by design, and that the current scope of the spec is just to focus on the syntax, I think we all agree on that. What I try to do is to look beyond what we have and try to share some critical views on the subject. I’m a supporter of the spec, did a lot of work to improve the specs while working with you on HiGHmed, and I think it can still be improved.
I’m clearly in favor of specifying syntax and semantics. As said: a lot of informal agreement which needs to be written down.
You can if you use the same schema to store the data, basically you need to borrow the archive from EHRBase and the AQL processing implementation in order for that to work. IMHO that has no point, just use EHRBase directly.
IMO EHRBase AQL is not unstable, is a fine piece of technology designed by very smart people. I personally respect that work a lot, it took a lot of effort to move EHRBase to the point it is now, so even if I work on competing products, EHRBase is an undeniable piece of engineering. Of course, it can still be improved (I even opened an issue on their repo about a bug I found last week). I can say the same about EHRServer an Atomik, those could also be improved, though they follow a very different design approach.
SAQM is not a general purpose querying language, so there is no concept of joining, since there are no tables. For aggregations, it just supports count.
The idea is that is a very simple way of querying the data you need, then you process the data you get as results. I believe in not delegating a lot of logic to the database, so complex processing is done by the system, not the database. That is one of our design principles.
Check the videos and you will see what I’m talking about.